100,99 €
Mehr erfahren.
- Herausgeber: John Wiley & Sons
- Kategorie: Wissenschaft und neue Technologien
- Sprache: Englisch
Presents the methodology and applications of ODE and PDE models within biomedical science and engineering
With an emphasis on the method of lines (MOL) for partial differential equation (PDE) numerical integration, Method of Lines PDE Analysis in Biomedical Science and Engineering demonstrates the use of numerical methods for the computer solution of PDEs as applied to biomedical science and engineering (BMSE). Written by a well-known researcher in the field, the book provides an introduction to basic numerical methods for initial/boundary value PDEs before moving on to specific BMSE applications of PDEs.
Featuring a straightforward approach, the book’s chapters follow a consistent and comprehensive format. First, each chapter begins by presenting the model as an ordinary differential equation (ODE)/PDE system, including the initial and boundary conditions. Next, the programming of the model equations is introduced through a series of R routines that primarily implement MOL for PDEs. Subsequently, the resulting numerical and graphical solution is discussed and interpreted with respect to the model equations. Finally, each chapter concludes with a review of the numerical algorithm performance, general observations and results, and possible extensions of the model. Method of Lines PDE Analysis in Biomedical Science and Engineering also includes:
- Examples of MOL analysis of PDEs, including BMSE applications in wave front resolution in chromatography, VEGF angiogenesis, thermographic tumor location, blood-tissue transport, two fluid and membrane mass transfer, artificial liver support system, cross diffusion epidemiology, oncolytic virotherapy, tumor cell density in glioblastomas, and variable grids
- Discussions on the use of R software, which facilitates immediate solutions to differential equation problems without having to first learn the basic concepts of numerical analysis for PDEs and the programming of PDE algorithms
- A companion website that provides source code for the R routines
Sie lesen das E-Book in den Legimi-Apps auf:
Seitenzahl: 358
Veröffentlichungsjahr: 2016
Ähnliche
Table of Contents
Cover
Title Page
Copyright
Dedication
Preface
About the Companion Website
Chapter 1: An Introduction to MOL Analysis of PDEs: Wave Front Resolution in Chromatography
1.1 1D 2-PDE model
1.2 MOL routines
1.3 Model output, single component chromatography
1.4 Multi component model
1.5 MOL routines
1.6 Model output, multi component chromatography
References
Chapter 2: Wave Front Resolution in Vegf Angiogenesis
2.1 1D 2-PDE model
2.2 MOL routines
2.3 Model output
2.4 Conclusions
References
Chapter 3: Thermographic Tumor Location
3.1 2D, 1-PDE model
3.2 MOL analysis
3.3 Model output
3.4 Summary and conclusions
References
Chapter 4: Blood-Tissue Transport
4.1 1D 2-PDE model
4.2 MOL routines
4.3 Model output
4.4 Model extensions
4.5 Conclusions and summary
References
Chapter 5: Two-Fluid/Membrane Model
5.1 2D, 3-PDE model
5.2 MOL analysis
5.3 Model output
5.4 Summary and conclusions
Chapter 6: Liver Support Systems
6.1 2-ODE patient model
6.2 Patient ODE model routines
6.3 Model output
6.4 8-PDE ALSS model
6.5 Patient-ALSS ODE/PDE model routines
6.6 Model output
6.7 Summary and conclusions
Appendix - Derivation of PDES for Membrane and Adsorption Units
A.6.1 PDEs FOR MEMBRANE UNITS
A.6.2 PDEs FOR ADSORPTION UNITS
References
Chapter 7: Cross Diffusion Epidemiology Model
7.1 2-PDE model
7.2 Model routines
7.3 Model output
7.4 Summary and conclusions
Reference
Chapter 8: Oncolytic Virotherapy
8.1 1D 4-PDE model
8.2 MOL routines
8.3 Model output
8.4 Summary and conclusions
Reference
Chapter 9: Tumor Cell Density in Glioblastomas
9.1 1D PDE model
9.2 MOL routines
9.3 Model output
9.4 -Refinement error analysis
9.5 Summary and conclusions
References
Chapter 10: Mol Analysis with a Variable Grid: Antigen-Antibody Binding Kinetics
10.1 ODE/PDE model
10.2 MOL routines
10.3 Model output
10.4 Summary and conclusions
Appendix: Variable Grid Analysis
References
Appendix A: Derivation of Convection-Diffusion-Reaction Partial Differential Equations
Reference
Appendix B: Functions dss012, dss004, dss020, vanl
Reference
Index
End User License Agreement
Pages
xi
xii
xiii
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
351
352
353
354
355
356
Guide
Cover
Table of Contents
Preface
Begin Reading
List of Illustrations
Chapter 1: An Introduction to MOL Analysis of PDEs: Wave Front Resolution in Chromatography
Figure 1.1 Diagram of a chromatographic column
Figure 1.2 Numerical solutions of eqs. (1.8),
ncase=2
,
ifd=3
, pulse BC
Figure 1.3 Numerical solution of eqs. (1.1b) , (1.2b) , ncase=2, ifd=1
Figure 1.4 Comparison of the numerical and analytical solutions of eqs. (1.1b) , (1.2b)
Figure 1.5 Numerical solution of eqs. (1.1b) , (1.2b) for ncase=2, ifd=2
Figure 1.6 Comparison of the numerical and analytical solutions of eqs. (1.1b) , (1.2b)
Figure 1.7 Numerical solution of eqs. (1.1b) , (1.2b) , ncase=2, ifd=3
Figure 1.8 Comparison of the numerical and analytical solutions of eqs. (1.1b) , (1.2b)
Figure 1.9 Numerical solution of eqs. (1.1b) , (1.2b) , ncase=2, ifd=4
Figure 1.10 Comparison of 5pbu (ifd=3) and van Leer (ifd=4)
Figure 1.11 Comparison of 5pbu (ifd=3) and van Leer (ifd=4)
Figure 1.12 Comparison of the numerical and analytical solutions of eqs. (1.1b), (1.2b) ncase=1, ifd=1, pulse BC
Figure 1.13 Numerical solution of eqs. (1.1b) , (1.2b) , ncase=2, ifd=1, pulse BC
Figure 1.14 Comparison of the numerical and analytical solutions of eqs. (1.1b) , (1.2b) ncase=1, ifd=2, pulse BC
Figure 1.15 Comparison of the numerical and analytical solutions of eqs. (1.1b) , (1.2b) ncase=1, ifd=3, pulse BC
Figure 1.16 Comparison of the numerical and analytical solutions of eqs. (1.1b) , (1.2b) ncase=1, ifd=4, pulse BC
Figure 1.17 Comparison of 5pbu (ifd=3) and van Leer (ifd=4), ncase=2
Figure 1.18 Comparison of 5pbu (ifd=3) and van Leer (ifd=4), ncase=2
Figure 1.19 Comparison of the numerical and analytical solutions of eqs. (1.8) ncase=1, ifd=3, pulse BC
Figure 1.20 Numerical solutions of eqs. (1.8), ncase=2, ifd=3, pulse BC
Chapter 2: Wave Front Resolution in Vegf Angiogenesis
Figure 2.1 Numerical and analytical solutions of eq. (2.1a), , (ncase=1)
Figure 2.2 Numerical and analytical solutions of eq. (2.1a), , (ncase=1)
Figure 2.3 Numerical solution of eq. (2.1a), ncase=2
Figure 2.4 Numerical solution of eq. (2.1b), ncase=2
Chapter 3: Thermographic Tumor Location
Figure 3.1 Solution to eqs. (3.1c), (3.2) and (3.3),
Figure 3.2 Solution to eqs. (3.1c), (3.2) and (3.3), ,
Chapter 4: Blood-Tissue Transport
Figure 4.1 Analytical and numerical solutions of eqs. (4.1), ifd=1,
Figure 4.2 Analytical and numerical solutions of eqs. (4.1),
ifd=2
,
Figure 4.3 Analytical and numerical solutions of eqs. (4.1),
ifd=3
,
Figure 4.4 Analytical and numerical solutions of eqs. (4.1),
ifd=4
,
Figure 4.5 Analytical and numerical solutions of eqs. (4.1), ifd=1,
Figure 4.4 Analytical and numerical solutions of eqs. (4.1), ifd=4,
Figure 4.8 Analytical and numerical solutions of eqs. (4.1), ifd=4,
Figure 4.7 Analytical and numerical solutions of eqs. (4.1), ifd=3,
Chapter 5: Two-Fluid/Membrane Model
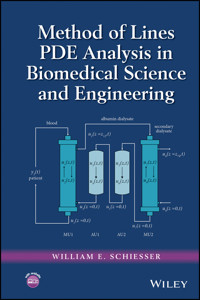
Figure 5.1 Diagram of a two-fluid/membrane system
Figure 5.2 Numerical solution
Figure 5.3 Numerical solution
Chapter 6: Liver Support Systems
Figure 6.1 ODE patient model
Figure 6.2 Solution of eqs. (6.2) and (6.3)
Figure 6.3 Artificial liver with four units
Figure 6.4a from eqs. (6.2) and (6.3)
Figure 6.4b from eqs. (6.4)
Figure 6.4c from eqs. (6.5), (6.6)
Figure 6.4d from eqs. (6.5), (6.6)
Figure 6.4e from eqs. (6.7)
Chapter 7: Cross Diffusion Epidemiology Model
Figure 7.1a (a) for ncase=2, (b) for ncase=2
Chapter 8: Oncolytic Virotherapy
Figure 8.1 vs with as a parameter for ncase=1
Figure 8.2 vs with as a parameter for ncase=2
Figure 8.3 vs with as a parameter for ncase=3
Figure 8.4 vs with as a parameter for ncase=4
Figure 8.5a vs with as a parameter for ncase=5
Figure 8.5b vs with as a parameter for ncase=5, nr=81
Figure 8.6 vs with as a parameter for ncase=6, nr=81
Figure 8.7a vs with as a parameter for ncase=7
Figure 8.7b vs with as a parameter for ncase=7
Figure 8.8a vs with as a parameter for ncase=8
Figure 8.8b vs with as a parameter for ncase=8 and a ramp
Figure 8.9a vs with as a parameter for ncase=9
Figure 8.9b vs with as a parameter for ncase=9
Chapter 9: Tumor Cell Density in Glioblastomas
Figure 9.1 (a) vs with , ncase=1, linear, (b) vs with , ncase=1, linear
Figure 9.2 (a) vs with , ncase=3, Gompertz, (b) vs with , ncase=3, Gompertz
Figure 9.3 (a) vs with , ncase=1, , (b) vs with , ncase=1,
Chapter 10: Mol Analysis with a Variable Grid: Antigen-Antibody Binding Kinetics
Figure 10.1 Schematic of diffusion-binding system
Figure 10.2a Uniform spatial grid
Figure 10.2d vs , uniform grid
Figure 10.2b Increment of uniform spatial grid
Figure 10.2c vs , uniform grid
Figure 10.3a Variable spatial grid
Figure 10.3d vs , variable grid
Figure 10.3b Incremental spacing of variable spatial grid
Figure 10.3c vs , variable grid
List of Tables
Chapter 1: An Introduction to MOL Analysis of PDEs: Wave Front Resolution in Chromatography
Table 1.1 Variables and parameters of eq. (1.1a)
Table 1.2 Selected numerical output for eqs. (1.1) to (1.4) from pde_1_main and pde_1 for ncase=1,2
Table 1.3 Numerical output for eqs. (1.1) to (1.4) for ncase=1,2, ifd=2
Table 1.4 Numerical output for eqs. (1.1) to (1.4) for ncase=1,2, ifd=3
Table 1.5 Numerical output for eqs. (1.1) to (1.4) for ncase=1,2, ifd=4
Table 1.6 Abbreviated comparison of output for eqs. (1.1) to (1.4) for ncase=2
Chapter 2: Wave Front Resolution in Vegf Angiogenesis
Table 2.1 Abbreviated Numerical Output at for ncase=1
Chapter 3: Thermographic Tumor Location
Table 3.1 Abbreviated Output from Listings 3.1 and 3.2
Table 3.2 Abbreviated Output from Listings 3.1 and 3.2 for
Chapter 4: Blood-Tissue Transport
Table 4.1 Numerical Output from the Main Program of Listing 4.2,
Table 4.2 Numerical Output from the Main Program of Listing 4.2,
Chapter 5: Two-Fluid/Membrane Model
Table 5.1 Indexing for the MOL analysis of eqs. (5.1)
Table 5.2 Numerical solution for eqs. (5.1) to (5.3)
Chapter 6: Liver Support Systems
Table 6.1 Numerical solution for eqs. (6.2) and (6.3)
Table 6.2 Numerical solution for eqs. (6.2) and (6.3) with eqs. (6.4) to (6.7) included
Chapter 7: Cross Diffusion Epidemiology Model
Table 7.1 Numerical values of the parameters in eqs. (7.1)
Table 7.2 Abbreviated output for ncase=1
Table 7.3 Abbreviated output for ncase=2
Table 7.4 Abbreviated output for ncase=3
Chapter 8: Oncolytic Virotherapy
Table 8.1 Abbreviated numerical output,
ncase=1
Table 8.2 Abbreviated numerical output, ncase=2
Table 8.3 Abbreviated numerical output, ncase=3
Table 8.4 Abbreviated numerical output, ncase=4
Table 8.5a Abbreviated numerical output, ncase=5
Table 8.5b Abbreviated numerical output, ncase=5, nr=81
Table 8.7 Abbreviated numerical output, ncase=6
Table 8.8 Abbreviated numerical output, ncase=7
Table 8.8a Abbreviated numerical output, ncase=8
Table 8.8b Abbreviated numerical output, ncase=8 and a ramp
Table 8.9 Abbreviated numerical output,
ncase=9
Chapter 9: Tumor Cell Density in Glioblastomas
Table 9.1 Abbreviated numerical output for ncase=2, logistic
Table 9.2 Abbreviated numerical output for ncase=3, Gompertz
Table 9.3 Abbreviated numerical output for ncase=1, linear
Chapter 10: Mol Analysis with a Variable Grid: Antigen-Antibody Binding Kinetics
Table 10.1 Dependent and independent variables of eqs. (10.1) and (10.4)
Table 10.2 Parameters and numerical values for eqs. (10.1) to (10.5)
Table 10.3 Abbreviated numerical output for the uniform grid
Table A.10.1 Prescribed ) pairs
Table 10.10 Abbreviated numerical output for ifcn=2, p=1.5
Method of Lines PDE Analysis in Biomedical Science and Engineering
WILLIAM E. SCHIESSER
Department of Chemical and Biomolecular EngineeringLehigh UniversityBethlehem, PA USA
Copyright © 2016 by John Wiley & Sons, Inc. All rights reserved
Published by John Wiley & Sons, Inc., Hoboken, New Jersey
Published simultaneously in Canada
No part of this publication may be reproduced, stored in a retrieval system, or transmitted in any form or by any means, electronic, mechanical, photocopying, recording, scanning, or otherwise, except as permitted under Section 107 or 108 of the 1976 United States Copyright Act, without either the prior written permission of the Publisher, or authorization through payment of the appropriate per-copy fee to the Copyright Clearance Center, Inc., 222 Rosewood Drive, Danvers, MA 01923, (978) 750-8400, fax (978) 750-4470, or on the web at www.copyright.com. Requests to the Publisher for permission should be addressed to the Permissions Department, John Wiley & Sons, Inc., 111 River Street, Hoboken, NJ 07030, (201) 748-6011, fax (201) 748-6008, or online at http://www.wiley.com/go/permissions.
Limit of Liability/Disclaimer of Warranty: While the publisher and author have used their best efforts in preparing this book, they make no representations or warranties with respect to the accuracy or completeness of the contents of this book and specifically disclaim any implied warranties of merchantability or fitness for a particular purpose. No warranty may be created or extended by sales representatives or written sales materials. The advice and strategies contained herein may not be suitable for your situation. You should consult with a professional where appropriate. Neither the publisher nor author shall be liable for any loss of profit or any other commercial damages, including but not limited to special, incidental, consequential, or other damages.
For general information on our other products and services or for technical support, please contact our Customer Care Department within the United States at (800) 762-2974, outside the United States at (317) 572-3993 or fax (317) 572-4002.
Wiley also publishes its books in a variety of electronic formats. Some content that appears in print may not be available in electronic formats. For more information about Wiley products, visit our web site at www.wiley.com.
Library of Congress Cataloging-in-Publication Data:
Names: Schiesser, W. E., author.
Title: Method of lines PDE analysis in biomedical science and engineering/
William E. Schiesser.
Description: Hoboken, New Jersey : John Wiley & Sons, 2016. | Includes
bibliographical references and index.
Identifiers: LCCN 2015043460 | ISBN 9781119130482
Subjects: LCSH: Differential equations, Partial–Numerical solutions. |
Numerical analysis–Computer programs.
Classification: LCC QA377 .S35 2016 | DDC 610.1/515353–dc23 LC record available at http://lccn.loc.gov/2015043460
To
Edward Amstutz
Glenn Christensen
Joseph Elgin
Harvey Neville
Preface
The reporting of differential equation models in biomedical science and engineering (BMSE) continues at a remarkable pace. In this book, recently reported models based on initial-boundary-value ordinary and partial differential equations (ODE/PDEs) are described in chapters that have the following general format:
1.
The model is stated as an ODE/PDE system, including the required initial conditions (ICs) and boundary conditions (BCs). The origin of PDEs based on mass conservation is discussed in Appendix A.
2.
The coding (programming) of the model equations is presented as a series of routines in R, which primarily implements the method of lines (MOL) for PDEs. Briefly, in the MOL,
The partial derivatives in the spatial (boundary value) independent variables are replaced by approximations such as finite differences, finite elements, finite volumes, or spectral representations. In the present discussion, finite differences (FDs) are used, although alternatives are easily included.
1
Derivatives with respect to an initial-value variable remain, which are expressed through a system of ODEs. An ODE solver (integrator) is then used to compute a numerical solution to the ODE/PDE system.
2
3.
The resulting numerical and graphical (plotted) solution is discussed and interpreted with respect to the model equations.
4.
The chapter concludes with a review of the numerical (MOL) algorithmperformance, general observations and results from the model, and possible extensions of the model.
The source of each model is included as one or more references. Generally, these are recent papers from the scientific and mathematics literature. Typically, a paper consists of some background discussion of the model, a statement of the model ODE/PDE system, presentation of a numerical solution of the model equations, and conclusions concerning the model and features of the solution.
What is missing in this format are the details of the numerical algorithms used to compute the reported solutions and the coding of the model equations. Also, the statement of the model is frequently incomplete such as missing equations, parameters, ICs, and BCs, so that reproduction of the solution with reasonable effort is virtually impossible.
We have attempted to address this situation by providing the source code of the routines, with a detailed explanation of the code, a few lines at a time. This approach includes a complete statement of the model (the computer will ensure this) and an explanation of how the numerical solutions are computed in enough detail that the reader can understand the numerical methods and coding and reproduce the solutions by executing the R routines (provided in a download).
In this way, we think that the formulation and use of the ODE/PDE models will be clear, including all of the mathematical details, so that readers can execute, then possibly experiment and extend, the models with reasonable effort. Finally, the intent of the detailed discussion is to explain the MOL formulation and methodology so that the reader can develop new ODE/PDE models and applications without becoming deeply involved in mathematics and computer programming.
In summary, the presentation is not as formal mathematics, for example, theorems and proofs. Rather, the presentation is by examples of recently reported ODE/PDE BMSE models including the details for computing numerical solutions, particularly documented source code. The author would welcome comments and suggestions for improvements ([email protected]).
William E. SchiesserBethlehem, PA, USAMay 1, 2016
1
Representative routines for the approximation of PDE spatial derivatives are listed in Appendix B.
2
Additional ODEs, which might, for example, be BCs for PDEs, can naturally be included in the model MOL solution. The solution of a mixed ODE/PDE system is demonstrated in some of the BMSE applications. Also, differential algebraic equations (DAEs) can easily be included in a MOL solution through the use of a modified ODE solver or a DAE solver.
About the Companion Website
This book is accompanied by a companion website:
www.wiley.com/go/Schiesser/PDE_Analysis
The website includes:
• Related R Routines